When dealing with copulas, R is a better option in my opinion, however, what could you do if you wish to use Python instead? There’s a good starting package called Copulalib which you can easily download here.
The package is really simple to use and very user-friendly I would say, it basically handles everything (pseudo-observations etc…) once you fed in the raw data. There is a simple example of implementation in the download page. Once the data has been fed into the function, the fitting is done automatically and the following parameters are generated:
-Spearman’s rho
-Kendall’s tau
-Theta (the parameter of the copula)
As of my understanding of the package, only Frank, Gumbel and Clayton copulas are available, and this of course could be a limitation, however it is for sure a good start. Another point which is problematic is that multidimensional copulas seem not to be supported.
Now for the real complaints: for some reason once the sample size is larger than 300 observations per variable (say 300 x and 300 y) the script raises an error saying that x and y must be of the same dimensions which is strange since they are already of the same size. Anyway maybe I did something incorrect.
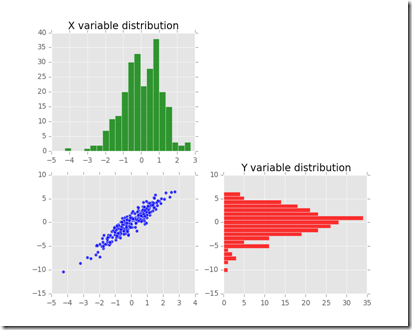
Here below is the short piece of code which generated the plots of the data and of the available copulas
Next I’m going to post a class for copulas.

Thanks for the nice post. I am also trying to move my R copula script to Python.
ReplyDeleteI agree that the current copulalib is quite limited, and I think that size greater than 300 problem is a bug.
Also wonder why the fitting procedure is not taking U and V values in [0,1] and instead taking raw data values. Suppose I have X and Y raw data and fitted in certain (but different) distributions that I preferred, I would want to use my own set of Us and Vs to fit the copula.
I am also wandering why the fitting procedure is taking the raw data values directly. Have you got any answer.
DeleteThank you!! I was looking just for this :)
ReplyDeletethank you very useful post.
ReplyDeleteI think the error mentioned in the post, related to the large size of x and y is due to the line 58 of copulalib.py. There is a conditional statement, comparing the sizes of X and Y, by using is not, instead of using !=. The is not operator is checking if the sizes are not the same object, rather then check if they have not the same value.
to avoid the error you can modify the following code in the copulalib.py (line 58):
before:
# input array should have same zie
if X.size is not Y.size:
raise ValueError('The size of both array should be same.')
# input array should have same zie
if X.size != Y.size:
raise ValueError('The size of both array should be same.')
Grateful,
Luca
Thanks for pointing the bug and solution. I have updated the package in pip.
DeleteThank you very much for this post. If I got correct here the intention is to create a dependence structure for U and V. The support of the numbers is 0,1. Inverting the random numbers using the desire analytical distribution should return a multivariate distribution. So let's suppose X and Y are lognormal, if I got correct I should invert respectively the distribution in U and V to get the multivariate. I tried to call generate_xy (not used in the code above but included in the library) but I'm getting back that the module 'statistics' has no attribute 'cpdf'. Btw, if I got correct, generate_xy is using a Kernel approach that can be replaced by the logic I explained above (inverting U and V using the lognormal). Could you please help me to understand if what above is correct?
ReplyDeleteBtw, I'm Andrea. Nice to talk to you. :-)
ReplyDelete