Another subject we took in the statistics class was the Gini index.
Gini index or ratio or coefficient is used to calculate how much a certain transferable phenomenon such as income or stocks for instance, is concentrated.
For example, say you are evaluating a company and you’d like to know more about how the shares are divided among the shareholders. You could use Gini index for that!
I’ve calculated the index using R and random data you can download here. In case you’d like to know more about Gini index check here.
Here my simple R implementation of the index.
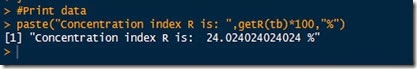
Here below are the results
It looks like the data I used shows a 24% concentration. Cool!


No comments:
Post a Comment